Drosera paleacea, a pygmy Drosera with the lowest number of chromosomes in the genus.
Chromosome number could be but is not considered a defining attribute of a taxonomic species. It is not a defining feature because taxonomic species are defined by characters that can be observed in dried herbarium specimens. Chromosome number is not an effective feature in the definition of biological species either although differences in chromosome number can produce different biological species. In spite of our lack of attention to chromosomes and chromosome numbers in formally defining species, it is the chromosomes and the genetic material they contain that define true species and control the evolution of new species.
If you already know about chromosomal evolution or do not care, jump directly to information on Drosera chromosome numbers.
There are very few references to the source material in the main discussion below. Please see the annotated bibliography at the bottom of this page for additional information.
What are chromosomes?
Chromosomes got their name by early cell biologists when they observed certain dark-staining objects in dividing cells. At the time they had no idea what these dark bodies were for or why they did the dance they observed. In human dividing cells 46 chromosomes can be observed. Drosera can have anywhere from 8 to 80 chromosomes depending on the species. It was not until the early 20th century that it was demonstrated that chromosomes contain the material of hereditary and it took until the middle 20th century to determine that DNA in chromosomes was the principle coding molecule of heredity. The heritable instructions for constructing an organism, or genes, are encoded in DNA and the whole set of instructions is called the genome. Each chromosome consists of one very long molecule of DNA twisted around proteins. The chromosomal proteins play a role in the regulation of DNA and are called the epigenome. This whole package of DNA (genome) and protein (epigenome) in a chromosome is what controls the development and maintenance of eukaryotic organisms (protists, fungi, animals, and plants).
At first glance this seems quite simple. We have a genome of instructions or genes that are like the instruction manual to build an organism and the epigenome which determines which instructions are used and when. As with any instruction manual, genetic instructions are not followed all at the same time. Correct timing of gene expression as controlled by the epigenome is critical to produce a functioning organism. Over generations, the genome and epigenome coevolve and change as an organism adapts to its environment. If this was the total story of life, life would be quite boring and we would not be here to be bored by it. Simply adapting to environments is not sufficient. Even if you add the fact that the "environment" is a moving target as the Earth wobbles making it warmer and cooler, continents move changing climates and sea level, and meteors and comets crash into the Earth, it is not enough perturbation to produce as many species and variety of life forms as we have today and have had in the past. There has to be something inherent in species or at least surviving species that produces the tremendous variation in life we see on this planet.
There are two factors related to chromosomes and species that work separately and together to make life interesting. The one that has been known for centuries is hybrids. Hybrids bring together genomes and epigenomes that have evolved in different directions and recombines them. The second factor has only been known since the middle of the 20th century but it was not until the 1980's that the full impact of transposable elements in the genome have been appreciated. Without hybridization and transposable elements we would not be here.
Chromosomes and hybrids.
The fate of plant hybrids in nature depends very much on the compatibility of the chromosomes of the parents, which parent is the seed parent, and the exact circumstances of the event. This process is totally independent of taxonomic characters used to define species. As long as the plants can physically interbreed and do they will form hybrids. The more similar the chromosomes (the genome and epigenome as well as chromosome number) the more likely hybrids will form and reproduce but also the more likely the net result will only be the introgression of genetic material from one species to the other. The more different the chromosomes of the hybrids are the more likely hybridization will result in a speciation event.
Introgression of characters via fertile hybrids
For smooth introgression to occur the parent species need to be closely related enough that the hybrids are fully fertile among themselves and with at least one of the parental species. The early generation hybrids probably would be physically recognizable but this condition is not stable. If the hybrids continue to interbreed with one or both of the parental populations then over time the hybrids would effectively be subsumed into a parental population possibly adding new characters. The only evidence of the event much later would be the presence of chromosomes or chromosome segments not found in other populations of the same species, possibly mitochondria or chloroplast variants not otherwise found in one parental species but found in the other parental species, and possibly some local variation in taxonomic traits. If some of the injected chromosomes have genes very advantageous to the species those chromosome elements can spread widely within the species very quickly erasing evidence of the event.
Without a full genome analysis it would be difficult to know if the variation found in a species exists because of genes that have been there all along, mutations that occurred in local populations, or genes injected via introgression from another species. Among carnivores related to Drosera, introgression has happened in Nepenthes in the past and is happening now. It can easily happen in Nepenthes because as far as we know all Nepenthes species and their hybrids are interfertile. In Drosera there may be something similar going on within the Drosera petiolaris-group of species and some of the African tetraploids but adequate studies of the situations have not been done or published.
Introgression of characters via almost sterile hybrids
If the chromosomes of hybrids are still close enough to produce a viable plant but sufficiently different that the plant is almost sterile we start getting into situations where it is difficult for the hybrids to be subsumed by the parents and more likely they will be able to become a new species. The hybrids with different chromosomes can grow and flower because the non-reproductive cells do not pair homologous chromosomes to divide. This kind of division is called mitosis. However cells destined to become pollen and ovules must pair their homologous chromosomes in the process we call meiosis. If the chromosomes can not pair then they will assort at random producing gametes with unusual combinations of chromosomes. This results in the plants being virtually sterile because the odds of compatible pollen and ovules getting together are very low.
There are other ways hybrids can be effectively sterile. Hybrids can be also be sterile if certain classes of transposable elements are not silenced in germ cells. The activity of the transposable elements can cause chromosome breaks and gene mutations that make it even less likely that proper pollen and ovules are formed. And hybrids can be sterile if there is a genetic imbalance in endosperm cells of the seed. Endosperm cells are triploid following the merger of two haploid cells from the seed parent and one haploid cell from the pollen parent. This ratio does not have to be exact but if it is not close enough the endosperm fails to form starving or drying the embryo. Drosera seeds may not have enough endosperm for this to be a major factor but there are many species where it is a serious factor in limiting hybridization.
A lot of things have to go right to produce viable seeds. When things do go right, almost sterile hybrids can inject new chromosomes into a host population in much the same way as fertile hybrids except at much lower frequency. This happens because with a very low probability, pollen from the hybrid will have exactly the complement of chromosomes as one of the parental species and will successfully produce back-cross individuals. It can have a greater effect on parent species than simple introgression because the introgressed chromosomes are from more distant relatives.
An example of introgression from an almost sterile hybrid can be seen in Senecio vulgaris, a non-carnivore in the daisy family. It is a tetraploid and native to the UK. The introduced diploid Senecio squalidus crossed with it to form an almost sterile triploid. The triploid produced pollen at a very low frequency that had the appropriate chromosome complement to produce a fertile back-cross to the tetraploid Senecio vulgaris. This resulted in the transfer of a flower characteristic that pollinators liked so much, the new introgression trait spread to other populations of the species. Generation of the triploid hybrid bridge was not necessary for this to happen but it did make it much more likely.
Hybrid speciation without change in chromosome numbers
The almost sterile hybrids can also result in the formation of a new species. New species can be formed in the same way as the introgression of Senecio squalidus traits into Senecio vulgaris except rather than a one or a few traits being conserved and the rest of the genome swamped out, a new species can form that has combined and unique traits. In a different occurrence of the same type of hybridization event between Senecio vulgaris and Senecio squalidus, the tetraploid hybrid was given the species name Senecio eboracensis because it is so different from the parents.
The above scenarios can happen when the range of closely related species abut or overlap or when long distance dispersal events occur. Many of the cases we know about involve range overlap because both parents are obvious and present. However long distance dispersal bringing one or a few plants of one species into the range of another can have the same effects only once the initial hybrids have gone the only evidence of what happened would be in the genomes of the descendents of the hybrids.
There is a huge amount of variation within Drosera spatulata. We do not know how much of this variation is simply local adaptations and how much of it results from introgression from very rare long distance dispersal of one type into the population of a different type.
Polyploidy and speciation
Hybridization and introgression between species happens all the time and is important for introducing new variants into populations and species. This "sharing" between species is important in the same way that "sharing" between humans has a use but is not very creative in the short term. What is really important evolutionarily is when something crazy happens which results in something totally new. This is what polyploidy does. In some ways you can argue polyploids are instant species.
Polyploidy or genome duplication has been an important factor in the evolution of life on this planet. Humans are diploidized octoploids. That is, the line to humans from the first vertebrates experienced two genome duplication events. Genome trimming and fertility selection makes our chromosomes appear to be a diploid complement but an analysis of our genome in comparison to the early vertebrate amphioxus clearly shows the evidence of duplications introduced by polyploidy. It has been proposed that amphioxus is itself a polyploid but the evidence for that is tenuous because the events happened so long ago too many pieces of the puzzle are missing.
The usual way genome duplications occur is via almost sterile hybrids between close but not too closely related species like we have seen above for introgression of characters and introgression speciation except in this case the hybrid produces something totally new on its own without backcrossing to one of the parents. That something new results from a complete genome duplication of the hybrid producing a fully fertile, allopolyploid species. There is another good example of this in Senecio as well as more examples in Tragopogon, Glycine, Nicotiana, and Brassica (see reference list below). They all basically say any time you have almost sterile homoploids or any odd-ploids (triploids, pentaploids, etc.) there will be neo species generated. And in all cases adequately studied it was found the events happened multiple times within and between populations producing an already diverse and evolution-ready species. We have three examples of this process in Drosera, each farther along the evolutionary landscape. The Drosera hybrids have not been studied as extensively as the examples in the reference list.
Drosera x hybrida
Drosera filiformis and Drosera intermedia are two closely related species found in North America. They both have a diploid complement of 2n=20 chromosomes. Drosera filiformis is found totally within the range of Drosera intermedia but the two do not normally produce hybrids in the wild. Part of the reason for not producing hybrids is they bloom at different times and tend to prefer slightly different habitats. Artificial hybrids between the two can be made and we know of three cases where hybrids occurred in nature. The homoploid, 2n=20, hybrids, named Drosera x hybrida, are virtually sterile presumably because the parental chromosomes can not pair properly in meiosis. At very low levels the hybrids have produced viable seeds that produced tetraploid plants, 2n=40. Homoploid/diploid plants produce tetraploid offspring when a pollen grain containing the total chromosome complement fertilizes an ovule also with the total chromosome complement. This results in plants with essentially the merged genomes of the parents. These tetraploid plants are fertile among themselves but should produce almost sterile offspring when backcrossed to the parent species. The tetraploid plants are fertile because doubling the chromosomes gives each of the parental chromosomes a pairing partner in meiosis.
Drosera tokaiensis
In Japan, the diploid, 2n=20, Drosera rotundifolia and tetraploid, 2n=40, Drosera spatulata overlap in range although they do have somewhat different habitat preferences. This is the southern end of the Drosera rotundifolia range and in Japan the species tends to be found in but is not limited to colder locations and sphagnum bogs. Japan is the northern limit of Drosera spatulata which tends to be found in the warmer winter regions along the Pacific coast. The hexaploid, 2n=60, Drosera tokaiensis, is found in a range of about 500 km in central Japan in the overlap region. Only one population with triploid, 2n=30, almost sterile Drosera spatulata by Drosera rotundifolia hybrids has been reported. Pentaploid hybrids between Drosera spatulata and Drosera tokaiensis, 2n=50, have also been reported. In nature Drosera tokaiensis is found growing with Drosera rotundifolia more often than Drosera spatulata but no natural hybrids between Drosera rotundifolia and Drosera tokaiensis have been reported in the literature. However there are hobbyist reports of aberrant plants that need to be studied further.
Chloroplast rbcL DNA sequences were determined for seven specimens of Drosera tokaiensis to establish the seed parent or parents of the species. All seven showed that Drosera spatulata was the seed parent. The initial triploid hybrids would have produced fully fertile allohexaploids by chromosomal nondisjunction in meiosis. It is not known how many times this occurred or the exact source populations for Drosera tokaiensis. Because the three species still live in such close proximity with each other, they can pass genes back and forth via the triploids and pentaploids. This is of little or no consequence to Drosera tokaiensis because it is already a mix of Drosera spatulata and Drosera rotundifolia genomes but it may have an influence on the Drosera spatulata and Drosera rotundifolia genomes in Japan.
Drosera anglica
Drosera anglica is a widespread tetraploid, 2n=40, species of known hybrid origin. Its parents are the diploid, 2n=20, Drosera rotundifolia and the diploid, 2n=20, Drosera linearis. Drosera anglica and Drosera rotundifolia are found in North America, Europe, and Asia while Drosera linearis is only found in North America in a narrow range from Montana to the Canadian Maritime provinces. Drosera rotundifolia and Drosera linearis form sterile, homoploid, 2n=20, hybrids where they grow together. As with Drosera x hybrida and triploid progenitor of Drosera tokaiensis, this homoploid hybrid will generate neo Drosera anglica at a very low rate. However it is obvious from its very extended distribution that Drosera anglica has been around for a long time. It would be interesting to study Drosera anglica across its full range to see if populations in different areas can be traced to different polyploidization events and to see the full extent that the peripheral populations have genomic differences from the populations with the neo individuals. Studies of the neo allopolyploid Tragopogon species have shown that those species had more than a dozen progenitors across numerous locations. But those populations and new species are too young to show what happens after an extended period of time as the genome(s) of the new species evolve. Drosera anglica is a good candidate for exploring genome evolution because it has populations with old and young lineages.
I said above that polyploidy would make exciting things happen evolutionarily. The creation of new species is interesting but Drosera tokaiensis and Drosera anglica are simply intermediates between their parents and ecologically they are essentially interchangeable with one or both of their parents. Drosera hybrida could be more interesting because it is ecologically intermediate between its parents. One reason it is not commonly found could be a lack of intermediate habitat. Drosera tokaiensis does tend to be a weed in captivity while its parents are not. But things could be more exciting than that. Drosera tokaiensis has the excuse of being a young species. Drosera anglica does not necessarily have that excuse. Part of the problem could be their chromosomes are too stable.
Undoubtably these Drosera species are experiencing diploidization of their genomes but it is happening in an excruciatingly slow way with apparently few side effects. In some hybrids and new species there is another factor that ramps up evolutionary change...
Chromosomes and transposable elements.
The second factor that makes life interesting on this planet is transposable elements. Transposable elements were discovered in the 1940's but it wasn't until whole genomes could be sequenced that the full importance of transposable elements could be seen. Almost half the human genome consists of transposable elements or their remains. This is not unusual. In plants transposable elements can consist of anywhere between the 10% of the Arabidopsis thaliana genome to the 85% of the Zea mays (maize or corn) and Hordeum vulgare (barley) genomes. And these numbers can be dramatically different between related species and even between clones within a species. Arabidopsis lyrata has three times the number of transposable elements in its genome compared to Arabidopsis thaliana. Zea mays clones can differ in transposable element composition by greater than 20%.
Generally in plants, the larger the genome, the higher the proportion of transposable element DNA. 60 megabases is a more than an adequate size genome to produce a complicated plant. Yet in plants the average genome size is one hundred times that size, 6,000 megabases, with more than half the genome being potentially active as well as defunct transposable elements.
There are two major classes of transposable elements. The most common class is the retrotransposons. Retrotransposons are related to retroviruses (HIV is a well known retrovirus) and are most likely derived from them. Retroviruses are known in plants but are not common. They are transmitted between plants by animals. Retroviruses can and do integrate into the genomes of the host (as HIV does) and can reproduce themselves at any time (as HIV will). Retrotransposons differ from retroviruses by not having the gene that produces the viroid capsule.
Retrotransposons have been called copy and paste transposable elements because they propagate in the genome by producing copies of themselves that in turn integrate somewhere else. The genes within the transposable element can get transcribed into RNA just like any other gene and proteins produced. One of those proteins is reverse transciptase that will transcribe the RNA transcript into DNA which can then be inserted into chromosomal DNA. These proteins can even help degenerate retrotransposons lacking any and all the genes to propagate within the genome. And instead of duplicating just retrotransposons, they can duplicate the region between closely spaced retrotransposons as well.
The other major class of transposable elements are DNA transposons. They are cut and paste transposable elements that rely on properties of DNA and a transposase enzyme to excise and insert themselves into chromosomes. DNA transposons will jump around the genome. Sometimes they leave pieces behind. Other times two transposons can excise together taking along a gene. This happened in Nepenthes where a chloroplast gene was duplicated to a chromosome in the nucleus or mitochondria. In this way as transposable elements move and insert into the genome they can cause changes in genes and gene regulation as well as move, duplicate, and delete genes.
With a genome infested with transposable elements, how can anything survive? The epigenome largely suppresses transposable elements. The transposable elements still move and cause damage but at a very low level. It is one of the trade-offs of life.
We would not be here without the massive genome restructuring facilitated by transposable elements. Evolutionarily, transposons are advantageous to species. They increase the speed of adaptation and the frequency of speciation. But they are deleterious to individuals. As well as the energetic load of maintaining and managing a genome full of jittery "junk", transposons can cause cellular disruptions like cancer and decrease fertility. This parallels the effects of polyploidy which may be good for the future of your species but is not necessarily good for the you.
Drosera rotundifolia chromosomes
There have not been any reports of transposon studies in Drosera and it may take a full genome analysis of a few species to determine the impact on transposons on Drosera evolution. However we do have the observation that Drosera rotundifolia and the species distal to it on the phylogenetic tree have larger chromosomes than Drosera spatulata and the species more proximal on the phylogenetic tree. This is not a polyploidy effect because even the diploid (2n=20) Drosera spatulata have tiny chromosomes, the same size as the tetraploids. Shirakawa, Hoshi and Kondo (2011) proposed that a likely reason for the larger chromosomes and probable larger genome size of Drosera rotundifolia is a transposable element outbreak in a progenitor of that species.
Polyploidization / genome trimming cycle.
Where transposable elements can get out of control and wreck havoc is in hybrids. Even in non-hybrids, mutant transposable elements can get out of control and the genome/epigenome needs to evolve to keep them in check. But in hybrids otherwise stable genomes can be combined resulting in transposable element outbreaks. Usually the outbreaks target one of the parental genomes, usually the one that is not the source of the transposons.
Hybrids mix genomes that can contain novel transposable elements or genome/epigenomes that have evolved in different directions to suppress transposable elements. This mixing of genomes can unbalance the suppression of transposable elements. Actually if the balance is too far out the hybrid can not survive in the first place. This is called hybrid dysgenesis. But in the case where the balance is sufficiently close what usually happens is nothing that can be easily detected. Even if the mutation rate is boosted by an order of magnitude or two that is still a low rate but significant in the long run. Remember that most plant species have been around for millions of years, unlike our own species.
Once transposons get into the genome the only way to get rid of them is via various non-typical recombination events. The transposons themselves present sites where those non-typical recombination events can occur. Also if chromosomes experience multiple breaks they may mis-repair because of the transposable elements being in many locations. Since polyploids have genes and whole chromosomes they can afford to lose, the result of non-typical recombination and mis-repair can be a genome revolution with massive losses and rearrangements of chromosomes. A good example of this in the Brassicaceae where a group of 6 species share a polyploidization event resulting in n=8 chromosomes. Two of the species have what appear to be the ancestral karyotype. The other 4 are essentially segmental aneuploids with a reduced number of chromosomes (n=5-7). They are missing chromosome segments including centromeres and have sequence inversions. One of these species, Arabidopsis thaliana, n=5, also happens to have one of the smaller genomes known for flowering plants and the lowest proportion of transposable elements in the genome. For Arabidopsis thaliana it is not known whether the transposable elements played a purely passive role in the genome trimming or if it was more active with the sites being targeted. What transposons are present are in locations with reduced recombination rates. The related species Arabidopsis lyrata with the ancestral n=8 chromosome complement has three times the number of transposable elements in its genome.
Some genome trimming happens because it can, but genome timing can be related to diploidization of the genome. New polyploids have twice the copy number of each gene. There are many genes that are strongly selected to only function at a certain level. Copies of those genes can be down-regulated, become non-functional, or even deleted to bring back the appropriate functional level. The presence and activity of transposable elements can speed this process.
Another factor driving genome trimming can be simply the nutrient and metabolic cost of maintaining so much chromosomal material in cells. Many carnivorous plants are nutrient limited more than typical plants and to the extent that cutting costs on unnecessary metabolic processes makes a difference, then eliminating extraneus chromosomal material will be of selective advantage.
New polyploids may also have trouble pairing certain chromosomes if the parent chromosomes are too similar. Rearrangement of chromosomal material can help promote proper segregation in meiosis. Again the presence of transposable elements could speed this process.
Polyploidization followed by genome trimming and diploidization has happened many times in the history of every plant species. The founder of the Droseraceae clade may have already been a diploidized tetraploid or hexaploid with x=6. This is based on genome analysis of other plants. When we have a full genome analysis of a few species in the Droseraceae we will know better how many times the genome duplication / genome trimming and diploidization cycle has occurred.
Holocentric chromosomes.
So far we have been looking at factors that affect chromosomes in general for all eukaryotes. They apply as much to humans as they do to Drosera. However Drosera chromosomes have a feature that is not common. Drosera chromosomes, as well as its closely related genera have diffuse or holocentric centromeres. Centromeres are where spindle fibers attach to chromosomes to separate them during meiosis and mitosis. Most eukaryotes have single centromeres however a few genera of eukaryotes have the centromeric function spread along the chromosome.
How do we know that Drosera has holocentric chromosomes? The first clue is the appearance of chromosomes during mitosis. Centromeric chromosomes have a constriction at the location of the centromere when viewed during mitosis. Here are some typical centromeric chromosomes from six different species:
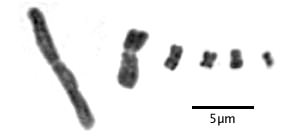
Typical centromeric chromosomes showing a constriction at the location of the centromere. The chromosome on the left is Drosophyllum lusitanicum; the others are from various non-carnivores. Images are details of one chromosome from full karyotypes in Rothfels and Heimburger (1968) and Hanson et al. (2003).
Notice the centromere is apparent on all the chromosomes although in species with tiny chromosomes it can be hard to see. You can also see the separate sister chromatids in the larger chromosomes.
Holocentric chromosomes during mitosis do not show the centromeric constriction. Notice also that in larger holocentric chromosomes the sister chromatids seem to be more widely separated than is seen in centromeric chromosomes:
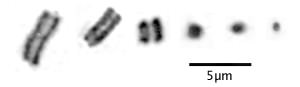
Typical Drosera chromosomes. Left to right: Drosera roseana, D. barbigera, D. walyunga, D. binata, D. rotundifolia, and D. capensis. Drosera roseana has among the largest Drosera chromosomes, Drosera capensis has among the smallest. Images are details of one chromosome from full karyotypes in Sheikh and Kondo (1995) and Rothfels and Heimburger (1968).
To see full karyotypes of Drosera regia, the only know Drosera species with centromeric chromosomes, and Drosera arcturi, with typical Drosera holocentric chromosomes, please see Shirakawa, Katsuya and Hoshi (2011) (PDF).
In larger holocentric chromosomes you can see a light band but it runs end-to-end separating the sister chromatids (duplicated chromosomes). In the smaller holocentric chromosomes, and in fact most Drosera chromosomes, with the best light microscopes you only see little blobs so we can not see this feature in all species. This uniform separation of sister chromatids indicates something fundamentally different is happening during chromosome segregation. The images are from cells treated with colchicine so there are no spindle fibers to orient the chromosomes or separate the sisters.
A second way to test if chromosomes are holocentric is to use x-radiation to break chromosomes in cells and see if the broken pieces can segregate normally during cell division. In centromeric chromosomes the broken pieces are generally lost. In holocentric chromosomes the broken pieces are usually conserved and are seen as microchromosomes. (In both cases rearrangements from reattachment of broken chromosomes in the wrong places are seen.) Sheikh, Kondo and Hoshi (1995) performed this test using Drosera dichrosepala and showed the chromosomes acted like holocentric chromosomes segregating microchromosomes.
How can it be that some genera and even certain species within a genus can have one kind of centromere and a closely related genus or group of species have another? For instance Drosera (except Drosera regia) and the closely related Aldrovanda appear to have holocentric chromosomes while Drosophyllum and apparently Dionaea have chromosomes with real centromeres. The answer is that even though it is possible to create a genome-based centromere with certain DNA sequences, animals and plants do not use fixed DNA-based centromere locations. Centromere function is determined by the epigenome. The epigenome decides where a centromere will be on a chromosome. Experiments have shown that if a chromosome lacks a centromere, the epigenome can create one. If a chromosome is constructed that should have two centromeres, the epigenome can disable one. Notice I say "can". There are studies that have shown this can happen but others that show what happens when it does not happen. Yes there are optimal locations for centromere function and yes in most eukaryotes those locations do not change much. The reason for that is centromeres and active gene locations are not compatible so centromeres tend to be in regions made up mostly of transposons and transposon remnants. Or to be more precise, transposons and transposon remnants accumulate where the centromere function hangs out.
The most studied plant genus with holocentric chromosomes is Carex (sedge). Carex is the poster child for counter examples in genetics. During meiosis in eukaryotes the homologous chromosomes pair and separate before the sister chromatids separate, but not in Carex. Carex does it the other way around and it appears Drosera does as well. If you are not a geneticist you might not be able to imagine how shocking this is. Meiosis is such a basic process and to have a few random genera in plants and animals segregating their chromosomes in opposite order is not at all expected. And there is more. In plants, including Drosera, pollen mother cells usually produce produce four pollen grains, but not in Carex. Carex only produces one pollen grain per mother cell. There are technical reasons for a plant species not to go there but Carex did. One of those reasons is meiotic drive. In meiosis where you have homologous chromosomes going separate ways and only one of those ways is into the future, there is always the possibility of one of the homologous chromosomes being on the right side of fate more often than not. Species commit suicide that way. That sounds a little over dramatic, and Carex is a little extreme in that it provides opportunities in both male and female meiosis, but it is true. In species like Nepenthes that have separate sexes and species like the pygmy, woolly, and tuberous Drosera that have flower self-incompatibility, meiotic drive can very quickly eliminate sexual reproduction if a chromosome can "cheat" in meiosis. There are theories that postulate that epigenetic control of chromosome function exists to keep the genome from cheating and holocentric chromosomes is the extreme case when the genome interferes with centromeric function and the epigenome reacts by dispersing the centromeres. I should say "appears" to react because what really happens is the populations and species with meiotic drive go extinct and the ones that counteract it survive.
For organisms with holocentric chromosomes it is believed that centromeric function occurs at a low level in the regions between genes all along the chromosomes. However it is not known if there are a handful of weak centromeres or if the spindle fibers can attach at a large number of locations. It is hard to study these chromosomes because they are hyper-condensed during meiosis and mitosis and so small you can not see details easily with a light microscope. But when you can see what is going on and are able to stain chromosomes differentially, in most genera with holocentric chromosomes it appears the spindle fibers "slide" down the chromosome and pull at an end. Either end will do. In Drosera the chromosomes move as if a small number of locations are being pulled sideways. The Aldrovanda chromosomes are too small to study so we do not know if they fit the Drosera pattern.
If Drosera holocentric chromosomes are not fully typical by having fewer and stronger dispersed centromeres and since centromeric magic is performed by the epigenome could there be an explanation for why some Drosera species-groups have a hypervariable number of chromosomes and others appear to have more stable numbers? Yoshikazu Hoshi observed that although all Drosera species he studied segregated their chromosomes in mitosis the same way, not all of them were the same in meiosis. Drosera petiolaris had typical holocentric chromosome movement in the first division of meiosis while Drosera rotundifolia chromosomes moved as if they only had one centromeric location. Hoshi proposed that Drosera rotundifolia and its related species have more stable chromosome numbers than Drosera petiolaris and its relatives because the non-Australian Drosera may be partly reverting back to a single centromere location, at least in the first meiotic division.
So why are holocentric chromosomes not the norm? There is a downside to them. Cells of organisms with centromeres that experience chromosome breaks that do not get repaired properly should not survive cell division. This is actually good because you want all your cells and especially those that become your offspring to be reading off the same script. However if the same thing happens in an organism with holocentric chromosomes, the broken pieces can assort properly in mitosis but all kinds of weirdness can happen in meiosis when the chromosomes fail to pair properly. In Carex, the chromosome numbers are all over the place, even within the same species. There are hints of chromosome number progressions from polyploidy that are typical for plants, but only hints. Most species are technically aneuploids and many species have chromosome number races without any difference in the total amount of genetic material. These extra chromosomes undoubtably are broken pieces. A lot of work has gone into studying Carex chromosomes but I think pygmy Drosera chromosomes are even more interesting and deserve more study.
Pygmy Drosera chromosomes.
Sheikh and Kondo published a comparison study of chromosomes in cells undergoing mitosis in 11 pygmy Drosera species with photos of each under different staining techniques. Across the 11 species they reported chromosome counts from 6 to 20 without any evidence of polyploidy. The more chromosomes a species had the shorter they were. So a species with 20 chromosomes like Drosera echinoblastus had about half medium chromosomes, and half small fragments while Drosera roseana had 4 very large and 2 large chromosomes. And to make matters worse, the Drosera roseana root tips cells they examined had counts of 2n=6, 7, and 8 with evidence of broken chromosomes. Drosera helodes also had cells with aberrant chromosomes. Unfortunately their study was done with mail order gemmae. What really needs to be done with the pygmy Drosera is studies like those done with Carex where populations were sampled to determine what is really happening in nature with respect to chromosomes rather then testing one or a few plants from clones that have been in cultivation. Kondo's results tell us something interesting is happening in this group but that just doing chromosome counts on one or a few clones from a whole species and just counting and measuring chromosomes is not sufficient to answer basic questions about the evolution of chromosomes in the pygmy Drosera.
Ploidy levels.
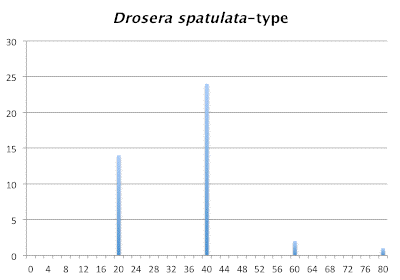
In species groups with well behaved chromosomes it is easy to determine the ploidy levels of each taxon. If you make a bar graph of chromosome numbers you will see a number progression like we see in the species derived from a Drosera spatulata-like ancestor. The progression is 20 : 40 : 60 : 80. We generally presume the lowest number is the diploid chromosome number. Of course if the number is greater than 2 the plant is a diploidized polyploid, but never mind. For all practical purposes 2n=20 would be considered diploid (2x) in this group. That would make 2n=40 tetraploid (4x), 2n=60 hexaploid, and so on. This can be confirmed if you do find 2n=30 and 2n=50 individuals and they are sterile. Odd-ploid (3x, 5x, etc.) plants are usually virtually sterile.
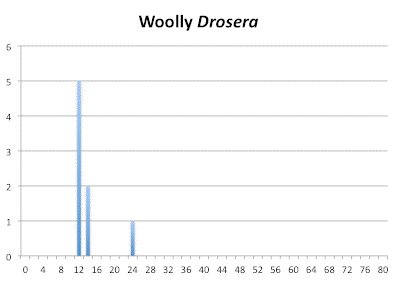
Within the woolly Drosera we find the progression (12,14) : 24. This would indicate diploid would be 2n=12 and tetraploid 2n=24. Notice this is about half the number of chromosomes for the diploid set compared to the Drosera spatulata-like ancestor and why I consider 2n=10 or 2n=12 to be ancestral for Drosera. If we look at the closest ancestors for Drosera spatulata we find the group of Australian species consisting of Drosera indica, Drosera hamiltonii, and the three Queensland sisters having 2n=28 and 2n=30. These have to be even-ploid because they are fertile but those numbers are odd-ploid in the Drosera spatulata-derived species. That means for them the base chromosome number is 2n=10 and thus they are hexaploid (6x). The 2n=28 for Drosera indica and Drosera hamiltonii the 2n=14 in the woolly Drosera are not bothersome in general because chromosomes can get lost or split following a polyploidization event. However the different numbers for the same species gives us a hint of trouble if the plants are correctly identified and are the same species.
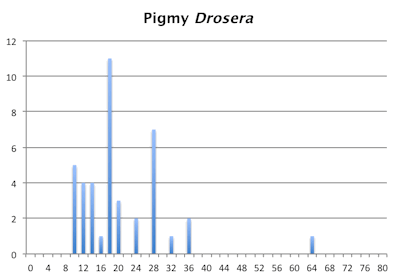
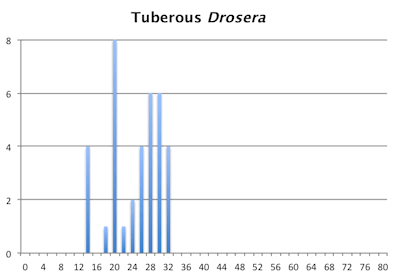
The bar graphs for chromosome number for the pygmy and tuberous Drosera are a mess. They are not as bad as the graph of chromosome numbers in Carex but still a mess. Within the pygmy and tuberous Drosera it appears most species are either tetraploids or hexaploids but that is assuming a low level of chromosome fragmentation. If they are all fragments then looking at chromosome numbers reverts to just a curiosity since we have no idea what we are looking at.
Phylogenies and chromosomes.
When we look at phylogenies what we are seeing is how a set of organisms is related through their genes. The rbcL phylogeny of Drosera is calculated from differences between the rbcL genes in the chloroplasts of each Drosera species. The branches in the tree follow the ancestry of the chloroplasts found in each species today; not the genome. Each branchpoint is when the chloroplast lineage split as determined by a change in its DNA sequence. Although this is not necessarily when the species lineage split it is a good approximation as long as hybridization is not involved. Hybridization violates the assumptions used to do the distance comparison and to draw the binary branching tree. A large study of Gossypium (cotton) showed that one fourth of the diploid Gossypium species they studied had evidence of introgression from genetically distant species as well as one lineage of five allotetraploid species that resulted from hybridization and polyploidy following a long distance dispersal event from Africa to the Americas. There is no way to see evidence for these events if plastid and nuclear DNA sequence data are combined and only one analysis and tree are produced.
Compare the locations of Drosera spatulata (Japan) and Drosera rotundifolia along with their child species Drosera tokaiensis and compare Drosera rotundifolia with its other child species Drosera anglica on the Drosera phylogeny page or the detail to the right. There is no indication in the rbcL phylogeny that Drosera tokaiensis and Drosera anglica contain a full Drosera rotundifolia genome. And by comparing the nuclear ribosomal ITS sequences we have no evidence among the sequences published that Drosera tokaiensis is related to Drosera spatulata. Since no sequences for Drosera linearis have been published we do not have any DNA evidence for the heritage of Drosera anglica. Yet we do know the heritage of those allopolyploids because the process is happening today and can draw a network relating the actual heritage of the species.
How many other species and lineages result from hybridization and polyploidy? If my estimates of ploidy level of species are correct then many key lineages are allopolyploids and thus hybrids. It is impossible to draw the true phylogeny using the data analysis techniques commonly used in molecular taxonomy because they can only draw a binary tree or some derivative of one when in fact the real phylogeny is a network. As was done in Gossypium, it is necessary to compare phylogenies based on individual nuclear genome and chloroplast DNA sequences and manually create a network of relationships. Some day we will be able to do this for more Drosera species.
-- John Brittnacher
November 2012
Definitions and (Links to Wikipedia) for more information.
DNA
Deoxyribonucleic acid. Contains the coded instructions for producing an organism. Part of a chromosome. (W DNA)
RNA
Ribonucleic acid. Among other functions can copy coded instructions from DNA for use in the manufacture of proteins. Can also interfere with the production of proteins and transposable element replication. (W RNA) (W RNAi)
Gene
The unit of heredity. It can refer to any part of the genome that contributes to an observable trait irrespective of the actual function of that DNA coding region. It can also refer to specific transcribed genomic regions and the adjacent regulatory region whether or not the function is known. (W Gene)
Protein
A chain of amino acids. Proteins are produced in cells from information encoded in DNA. (W Genetic Code) (W Protein)
Genome
The set of all the chromosomal DNA. Can also refer to the DNA sequence. The human genome is over 3 billion DNA bases long. The range in flowering plants is 63 million to 150 billion DNA bases long. (W Genome)
Epigenome
Chromosome proteins such as histones that package DNA and control gene expression. Epigenome control is stable through mitosis. During meiosis epigenomic programming may be lost except at centromeres. (W Histones)
Chromosome
The package of DNA and proteins containing the genome. (W Chromosome) (W Chromatin)
Centromere
A visible constriction on chromosomes. Spindle fibers attach to the centromeric histones to pull the chromosomes apart during cell division. In most species, chromosomes or chromosome fragments without centromeres are lost. (W Centromere)
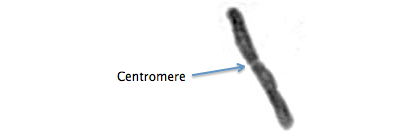
Drosophyllum lusitanicum chromosome. Detail from Rothfels and Heimburger (1968).
Holocentric Chromosome
Some species do not have discrete centromeres but instead have dispersed centromere function across the chromosomes. No constrictions can be seen during mitosis and meiosis and chromosome fragments can assort normally.
Mitosis
The part of normal cell division where the chromosomes are divided into the two daughter cells. (W Mitosis)
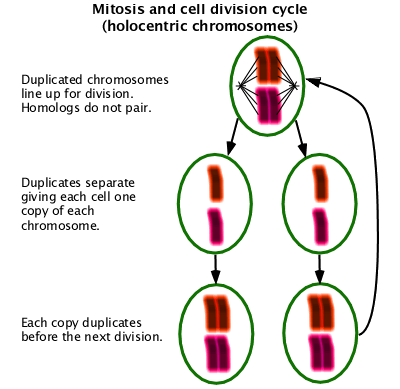
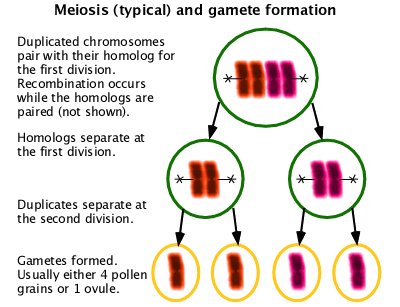
Meiosis
A special form of cell division used in gametogenesis with two cell division cycles resulting in four cells containing half the normal chromosome content. Unlike in mitosis, homologous chromosomes must pair in order to assort properly. In organisms with centromeric chromosomes, homologous chromosomes separate into separate cells at the first division. In organisms with holocentric chromosomes, homologous chromosomes assort at the second division. Usually recombination occurs as part of the pairing process. (W Meiosis)
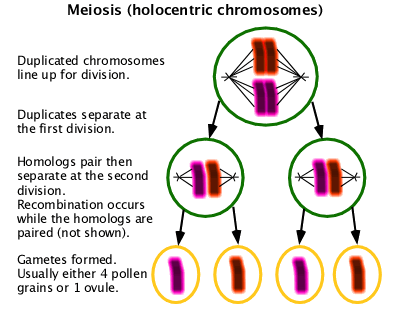
Karyotype
The account of the number and appearance of the set of chromosomes in an organism. (W Karyotype) (W Cytogenetics)
x
The basic chromosome number. The number of different chromosomes.
n
The number of chromosome pairs in mitosis. 2n chromosomes would be seen in mitotic cells.
Diploid
Containing two each of the basic chromosomes. 2n is the diploid chromosome number. In diploids, x = n.
Polyploid
Containing multiple diploid sets of chromosomes.
Tetraploid
Containing four of each chromosome, usually two diploid sets of chromosomes. 4x.
Triploid
Containing three of each chromosome, usual half of a diploid set and half of a tetraploid set. Usually almost sterile. 3x.
Triploid Bridge
Also homoploid or other odd-ploid bridge. Odd-ploid hybrids at a very low level produce gametes that facilitate chromosome transfers between species.
Other Common Ploidies
5x = pentaploid
6x = hexaploid
8x = octoploid
10x = decaploid
12x = duodecaploid
16x = hexadacaploid
Autopolyploid, Autotetraploid
Containing multiple sets of chromosomes from the same species. Autotetraploids contain two diploid sets. Higher ploidies are possible. Autopolyploids are of little evolutionary significance.
Allopolyploid, Allotetraploid, Allohexaploid, etc.
Containing multiple sets of chromosomes from different parent species. Allotetraploids have two different diploid chromosome complements; allohexaploids have a diploid and tetraploid complement, etc.
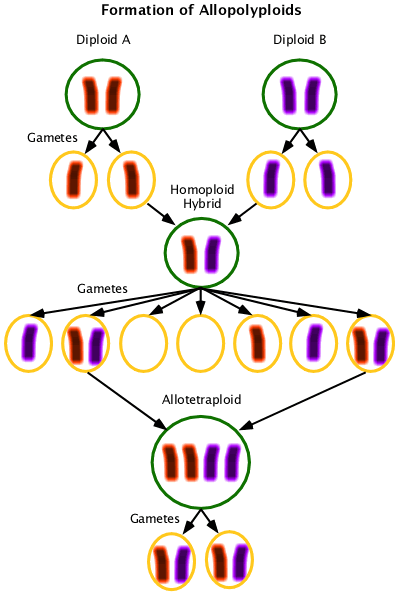
Allotetraploids are formed from interspecific hybrids that are not able to pair the chromosomes they received from their parents. At a low frequency these homoploids will produce unreduced gametes and at the square of that frequency those gametes will meet and form allotetraploids. Allotetraploids are fertile because they now have chromosomes that pair in meiosis. (This example ignores the sister chromatids (duplicate chromosomes) for simplicity.)

Drosera spatulata x rotundifolia hybrid. The plant is an almost sterile triploid homoploid since the parents are of different ploidy levels. Cross and photo by Ivan Snyder.

Drosera tokaiensis is a fertile allopolyploid with Drosera spatulata and Drosera rotundifolia as parents.

Drosera x hybrida tetraploid, a natural allotetraploid hybrid between Drosera intermedia and Drosera filiformis.
Neopolyploid, Synthetic Polyploid
Artificially produced polyploid.

Neo Drosera tokaiensis. The Drosera spatulata parent had pink flowers. Plant and photo by Ivan Snyder.

An artificial allotetraploid Drosera eloisiana 'Dr. Frankensnyder's Monster'. It was produced by Ivan Snyder from a sterile hybrid between Drosera intermedia and Drosera rotundifolia by treating leaf cuttings with colchicine.
Homoploid
A hybrid containing the same number of chromosomes as the parent species, half from each.

Drosera x hybrida. A natural homoploid hybrid between Drosera intermedia and Drosera filiformis.

Drosera x eloisiana 'Nightmare'. A natural homoploid hybrid between Drosera intermedia and Drosera rotundifolia.
Amphiploid
A hybrid containing both full chromosome sets from the parents. This would generally be double the homoploid. The same as an allopolyploid.
Aneuploid
Having the presence of extra chromosomes or missing chromosomes. Any deviation from the expected number of chromosomes. This can be from chromosome fusions or fissions or having extra chromosomes or missing chromosomes.
Segmental Aneuploid
Having chromosomes with extra, moved, or missing segments. A general term used when the exact nature of an unusual karyotype feature is not known or is unspecified.
Diploidization
The process where a polyploid for all practical purposes acts like a diploid. The term may refer to just the appearance of chromosomes in meiosis or may refer to the polyploid genome acting as or becoming functionally diploid.
Transposable Elements
Organized DNA sequences that can replicate and integrate into the genome. They are essentially DNA parasites that have played a major role in evolution. Also known as transposons. (W Transposable Elements)
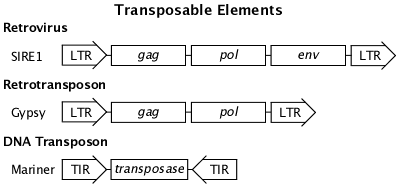
LTR = Long Terminal DNA sequence Repeats
TIR = Terminal Inverted DNA sequence Repeats
gag = produces multiple structural proteins
pol = produces reverse transcriptase, integrase, and protease
env = produces viral envelope proteins
transposase = enzyme that facilitates excision and insertion
Retrotransposon
Retrotransposons are related to and probably descended from retroviruses. Retrotransposons duplicate themselves in the genome by a copy and paste mechanism. They do not excise from the genome and can only be removed by deletion or unequal recombination. Insertion of retrotransposons can cause gene mutations.
The Zea mays (maize or corn) has in excess of 300,000 copies of retrotransposons in various families in its genome, Arabidopsis thaliana has on the order of 50 copies in its genome. The average plant species is closer to Zea mays than to Arabidopsis thaliana.
DNA Transposon
Transposons can move about the genome via a cut and paste mechanism facilitated by terminal inverted repeats that form DNA loops and the transposase enzyme. Movement of DNA transposons can cause gene mutations.
The Zea mays (maize or corn) has in more than 6,000 copies of DNA transposons in various families in its genome, Arabidopsis thaliana has 5 copies in two families in its genome.
Distribution of chromosome numbers within Drosera species groups.
Number of Chromosomes
Number of Chromosomes
Number of Chromosomes
Number of Chromosomes
Distribution of chromosome numbers in Drosera species groups. The Drosera spatulata-type includes all species derived from a plant similar to Drosera spatulata based on the Drosera rbcL phylogeny. In the pygmy Drosera graph, some counts are excluded because the same plant gave different numbers.
Bifurcating phylogenies can not tell the whole truth.
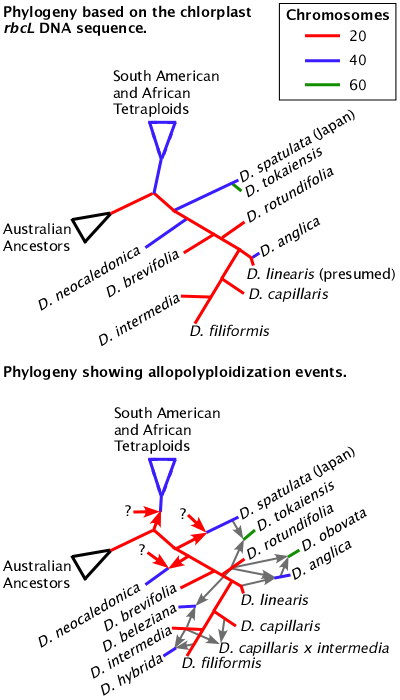
Phylogenies based on chloroplast (rbcL) and mitochondria DNA sequences are not able to show the true relationships between species if hybridization and allopolyploidy occur. Even for chromosomal genes, the standard analysis programs for phylogenies are not able to properly determine and graph the relationships between hybrids and allopolyploids. Hexaploid Drosera obovata is not known in nature but can be constructed from wild plants. Eventually this figure will be even more complicated when Drosera oblanceolata, Drosera ultramafica, and the various forms of Drosera spatulata are added.
Chromosome numbers reported for Drosera species.
Ploidy levels in the table are estimates based on x = 5, 6, or 7 except for the modern Drosera spatulata-type derived species that appear to be diploidized to x = 10. The ploidy shown for each species is based on the assumption that the species does not have a fragmented chromosome set. It will require full genome sequencing to determine actual ploidy level.
Note: (1) some counts are wrong and some source plants were incorrectly identified
(2) many of the counts are not properly published with details of methods and voucher specimens
(3) the ploidy levels shown are best guess and not based on an actual scientific measurement
| Ploidy levels for the basal Drosera are proposed according to the assumption that the ancestor of Drosera and the other genera in the Droseraceae and Nepenthaceae was a diploid (2x) with 2n=10, 12, or 14. |
|||||
| Species | Continent | 2n | Ploidy (x=5, 6, 7) |
Publication Authors*, Notes. |
|
| Drosera regia | AF | 34 | 6x | Behre | |
| Drosera arcturi | AU | 20 | 4x | Kondo & Whitehead | |
| Drosera glanduligera | AU | 22 | 4x | Kondo & Olivier | |
| Drosera sessilifolia | SA | 20 | 4x | Futagawa, Suzuki & K.Kondo, Rivadavia | |
| Drosera burmannii | AU, AS | 20 | 4x | Narasimhachar | |
| Drosera binata | AU | 32, 46, 64 | 6x, 8x, 12x | Behre:32, Sato:46, Kress:64 | |
| Drosera banksii | AU | 12 | 2x | Kondo & Lavarack | |
| Drosera petiolaris | AU | 12, 14 | 2x | Kondo:12, Hoshi & Kondo:14 | |
| Drosera dilatatopetiolaris | AU | 12 | 2x | Kondo | |
| Drosera falconeri | AU | 12 | 2x | Kondo | |
| Drosera ordensis | AU | 24 | 4x | Hoshi & Kondo | |
| Drosera lanata | AU | 12, 14 | 2x | Hoshi & Kondo:12,14 | |
| Drosera hamiltonii | AU | 28 | 6x | Kondo | |
| Drosera indica | AS | 28 | 6x | Venkatasubban | |
| Drosera adelae | AU | 28, 30 | 6x | Kondo:28, Kondo & Olivier:30 | |
| Drosera prolifera | AU | 30 | 6x | Kondo & Lavarack | |
| Drosera schizandra | AU | 30 | 6x | Kondo & Olivier | |
|
Ploidy levels for the pygmy Drosera are proposed according to the following assumptions: The known |
|||||
| Species | Continent | 2n | Ploidy (x=5, 6, 7) |
Publication Authors*, Notes. |
|
| Drosera allantostigma | AU | 28 | 4x | Lowrie(Conran) | |
| Drosera androsacea | AU | 20 | 4x | Lowrie(James) | |
| Drosera barbigera | AU | 10, 12 | 2x | Sheikh & Kondo:10, Lowrie(James):12 | |
| Drosera callistos | AU | 24 | 4x | Lowrie(James) | |
| Drosera citrina | AU | 18 | 4x | Lowrie(James) | |
| Drosera coomallo | AU | 36 | 8x | Lowrie(James) | |
| Drosera dichrosepala | AU | 18 | 4x | Kondo, Segawa & Nehira | |
| Drosera echinoblastus | AU | 20 | 2x | Sheikh & Kondo | |
| Drosera eneabba | AU | 14, 18 | 2x | Chen(James,Chen,Lowrie&Marchant):14, Lowrie(James):18 | |
| Drosera helodes | AU | 18 | 4x | Lowrie(James) | |
| Drosera hyperostigma | AU | 10 | 2x | Lowrie(James) | |
| Drosera lasiantha | AU | 24 | 4x | Lowrie(James) | |
| Drosera leioblasta | AU | 10 | 2x | Sheikh & Kondo, Lowrie(James) | |
| Drosera leucoblasta | AU | 14 | 2x | Kondo & Lavarack, Lowrie(James) | |
| Drosera leucostigma | AU | 28 | 4x | Lowrie(Conran) | |
| Drosera mannii | AU | 14 | 2x | Sheikh & Kondo, Lowrie(James), Chen(James,Chen,Lowrie&Marchant) | |
| Drosera meristocaulis | SA | ~32-36 | 6x or 8x | Rivadavia, Miranda, Hoogenstrijd, Pinheiro, Heubl & Fleischmann | |
| Drosera micrantha | AU | 10 | 2x | Lowrie(James) | |
| Drosera miniata | AU | ~32 | 6x | Lowrie(James&Lowrie) | |
| Drosera nitidula | AU | 28 | 4x | Kondo & Olivier, Lowrie(James) | |
| Drosera nivea | AU | 18 | 4x | Lowrie(James): n=9+0-2B | |
| Drosera occidentalis | AU | 28 | 4x | Lowrie(James) | |
| Drosera omissa | AU | 28 | 4x | Lowrie(James) | |
| Drosera oreopodion | AU | 14 | 2x | Sheikh & Kondo:14, Lowrie(James): n=5+3 | |
| Drosera paleacea | AU | 10 | 2x | Kondo & Lavarack | |
| Drosera patens | AU | 28 | 4x | Lowrie(James&Conran) | |
| Drosera pulchella | AU | 18 | 4x | Kondo, Segawa & Nehira, Lowrie(James) | |
| Drosera pygmaea | AU | 20, 28, 64 | 4x | Heitz:20(-22),?30, Kondo, Segawa & Nehira:28, Behre:(28-34),?32,64 | |
| Drosera roseana | AU | 12 | 2x | Lowrie(James):12, Sheikh & Kondo:6,7,8 | |
| Drosera scorpioides | AU | 18 | 4x | Lowrie(James) | |
| Drosera sewellae | AU | 18 | 4x | Sheikh & Kondo, Lowrie(James&Lowrie,n=18 is a typo) | |
| Drosera silvicola | AU | 12 | 2x | Lowrie(James) | |
| Drosera spilos | AU | 18 | 4x | Lowrie(James) | |
| Drosera stelliflora | AU | 18 | 4x | Lowrie(James) | |
| Drosera verrucata | AU | 12, 18 | 2x | Sheikh & Kondo:12, Lowrie(James): n=9+1B | |
| Drosera walyunga | AU | 16 | Sheikh & Kondo | ||
| Ploidy levels for the tuberous Drosera are proposed according to the following assumptions: The known diploids are all 2n=14. With tetraploids having 2n=20, 22, 24, 26, and 28, we assume diploids were 2n=10, 12, and 14. Hexaploids would then be 2n=30, 32, 34, 36, 38, 40, 42. |
|||||
| Species | Continent | 2n | Ploidy (x=5, 6, 7) |
Publication Authors*, Notes. |
|
| Drosera bulbosa | AU | 24 | 4x | Lowrie(James), Chen(James,Chen,Lowrie&Marchant) | |
| Drosera collina | AU | 20 | 4x | Lowrie(James) | |
| Drosera erythrorhiza | AU | 20 | 4x | Kondo & Lavarack, Lowrie(James) | |
| Drosera lowriei | AU | 14 | 2x | Lowrie(James) | |
| Drosera macrophylla | AU | 24 | 4x | Kondo & Lavarack | |
| Drosera major | AU | 20, 22 | 4x | Chen(James,Chen,Lowrie&Marchant):20, Lowrie(James):22 | |
| Drosera orbiculata | AU | 26 | 4x | Chen(James,Chen,Lowrie&Marchant) | |
| Drosera rosulata | AU | 26 | 4x | Chen(James,Chen,Lowrie&Marchant) | |
| Drosera tubaestylis | AU | 28 | 4x | Chen(James,Chen,Lowrie&Marchant) | |
| Drosera whittakeri | AU | 28 | 4x | Kondo & Lavarack | |
| Drosera basifolia | AU | 30 | 6x | Lowrie(James) | |
| Drosera bulbigena | AU | 14 | 2x | Lowrie(James) | |
| Drosera calycina | AU | 30 | 6x | Lowrie(James) | |
| Drosera gigantea | AU | 18, 28, 32 | 4x, 6x | Lowrie(James):18, Kondo:28, Chen(James,Chen,Lowrie&Marchant):32 | |
| Drosera graniticola | AU | 28 | 4x | Lowrie(James) | |
| Drosera heterophylla | AU | 30 | 6x | Kondo & Lavarack, Lowrie(James) | |
| Drosera heugelii | AU | 28 | 4x | Lowrie(James) | |
| Drosera menziesii | AU | 26, 30 | 4x, 6x | Kondo:26, Chen(James,Chen,Lowrie&Marchant):30 | |
| Drosera neesii | AU | 28 | 4x | Kondo & Lavarack | |
| Drosera radicans | AU | 14 | 2x | Chen(James,Chen,Lowrie&Marchant) | |
| Drosera stricticaulis | AU | 20 | 4x | Lowrie(Conran) | |
| Drosera humilis | AU | 20 | 4x | Lowrie(James) | |
| Drosera porrecta | AU | 20 | 4x | Chen(James,Chen,Lowrie&Marchant) | |
| Drosera purpurascens | AU | 20 | 4x | Chen(James,Chen,Lowrie&Marchant) | |
| Drosera ramellosa | AU | 26 | 4x | Kondo & Lavarack | |
| Drosera rupicola | AU | 20 | 4x | Chen(James,Chen,Lowrie&Marchant) | |
| Drosera auriculata | AU | 32 | 6x | Kress | |
| Drosera bicolor | AU | 32 | 6x | Lowrie(James) | |
| Drosera peltata | AU, AS | 32 | 6x | Kondo | |
| Drosera macrantha | AU | 30 | 6x | Chen(James,Chen,Lowrie&Marchant) | |
| Drosera pallida | AU | 30 | 6x | Lowrie(Chen&James) | |
| Drosera planchonii | AU | 30 | 6x | Kondo & Lavarack | |
| The ploidy levels for the Drosera descended from a Drosera spatulata-like species appear to be diploidized to 2n=20. Tetraploids (4x) are 2n=40, hexaploids (6x) are 2n=60, and octoploids (8x) are 2n=80. |
|||||
| Species | Continent | 2n | Ploidy (x= 10) |
Publication Authors*, Notes | |
| Drosera spatulata | AU, AS | 20, 40 | 2x, 4x | Rattenbury:20, Kondo:40, 4x hybrid origin? | |
| Drosera tokaiensis | AS | 60 | 6x | Kondo, hybrid origin: D. rotundifolia x D. spatulata | |
| Drosera neocaledonica | AS | 40 | 4x | Kondo, hybrid origin? | |
| Drosera brevifolia | NA, SA | 20 | 2x | Wood, Rivadavia | |
| Drosera linearis | NA | 20 | 2x | Wood | |
| Drosera rotundifolia | NA, EU, AS | 20 | 2x | Rosenberg | |
| Drosera anglica | NA, EU, AS | 40 | 4x | Rosenberg, hybrid origin: D. rotundifolia x D. linearis | |
| Drosera filiformis | NA | 20 | 2x | Levine, Rivadavia | |
| Drosera tracyi | NA | 20 | 2x | Wood | |
| Drosera capillaris | NA, SA | 20 | 2x | Wood | |
| Drosera cayennensis | SA | 20 | 2x | Rivadavia | |
| Drosera communis | SA | 20 | 2x | Rivadavia | |
| Drosera viridis | SA | 20 | 2x | Rivadavia | |
| Drosera intermedia | SA, NA, EU | 20 | 2x | Rosenberg:20, Rivadavia:20, (Rogers:40 = D. anglica?) | |
| Drosera roraimae | SA | 20 | 2x | Rivadavia | |
| Drosera hirtella | SA | 20 | 2x | Rivadavia | |
| Drosera hirtella var. lutescens | SA | 20 | 2x | Rivadavia | |
| Drosera ascendens | SA | 40 | 4x | Rivadavia | |
| Drosera camporupestris | SA | 40 | 4x | Rivadavia | |
| Drosera chrysolepis | SA | 40 | 4x | Rivadavia | |
| Drosera graminifolia | SA | 40 | 4x | Futagawa, Suzuki & K.Kondo, Rivadavia | |
| Drosera grantsaui | SA | 40 | 4x | Rivadavia | |
| Drosera graomogolensis | SA | 40 | 4x | Rivadavia | |
| Drosera montana | SA | 40 | 4x | Hoshi & Kondo | |
| Drosera tentaculata | SA | 40 | 4x | Rivadavia | |
| Drosera tomentosa | SA | 40 | 4x | Rivadavia | |
| Drosera villosa | SA | 40 | 4x | Futagawa, Suzuki & K.Kondo | |
| Drosera aliciae | AF | 40, 80 | 4x, 8x | Rivadavia:40 ,Behre:80, 8x hybrid origin? | |
| Drosera capensis | AF | 40 | 4x | Behre, Rivadavia | |
| Drosera cuneifolia | AF | 40 | 4x | Debbert | |
| Drosera venusta | AF | 40 | 4x | Debbert | |
| Drosera dielsiana | AF | 40 | 4x | Hoshi & K.Kondo | |
| Drosera madagascariensis | AF | 40 | 4x | Kress | |
| Drosera collinsiae | AF | 40 | 4x | Nicholas & K.Kondo | |
| Drosera burkeana | AF | 20 | 2x | Kondo, this needs to be verified | |
| Drosera trinervia | AF | 40 | 4x | Hoshi & Kondo | |
| Drosera hilaris | AF | 40 | 4x | Hoshi & Kondo | |
| Drosera cistiflora | AF | 40, 60 | 4x, 6x | Kondo & Olivier:40, Behre:60, Rivadavia:60, 6x hybrid origin? | |
| Drosera pauciflora | AF | 40 | 4x | Hoshi & Kondo | |
|
* Some publications have previously unpublished data by others: |
|||||
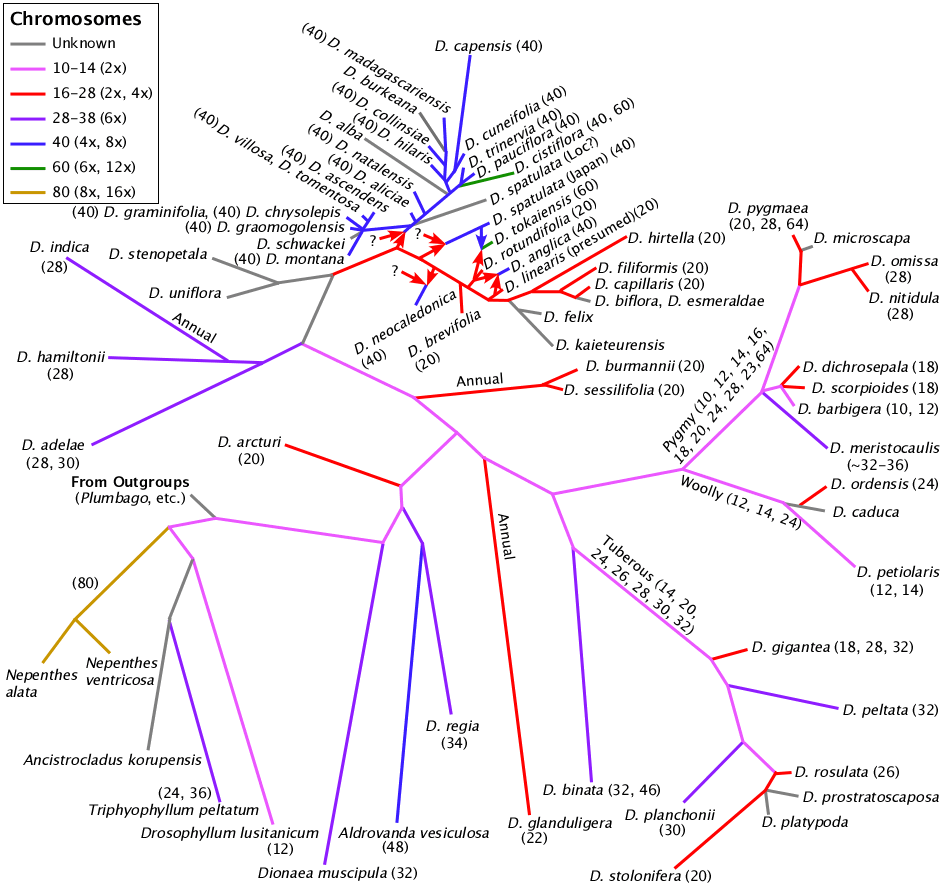
Chromosome numbers displayed on a modified rbcL Drosera phylogeny.
The rbcL phylogeny is based on a chloroplast gene and thus can not show the full complexity of the Drosera phylogeny owing to the occurrences of allopolyploidy. Drosera tokaiensis and Drosera anglica are allopolyploids where we know the parent species. To show them correctly requires deviating from the binary tree usually drawn for phylogenies. Drosera spatulata and Drosera neocaledonica are allopolyploids where we do not know the parents. The tetraploid South American and African species also result via one or more polyploidy events. There are other locations on the phylogeny where allopolyploidy is likely however at this point we can not confirm the details.
The choice for the chromosome numbers (colors) assigned to the interconnecting branches uses the same assumptions as the choice for ploidy level in the table above. The backbone on the branch leading from Drosera arcturi to Drosera adelae is shown as 10 or 12 because Drosera adelae appears to be a hexaploid. Hexaploids usually are derived from a tetraploid and a diploid. The tree can only show one of those.
For more information please see:
Drosera chromosomes and phylogeny
Kondo, Katsuhiko (1969) Chromosome Numbers of Carnivorous Plants. Bulletin of the Torrey Botanical Club 96(3): 322-328. (jstor)
Rothfels, K. and M. Heimburger (1968) Chromosome size and DNA values in sundews (Droseraceae). Chromosoma 25:96-103.
Kondo, K. and B. Whitehead (1971) Chromosome number of Drosera arcturi Hook. The Journal of Japanese Botany, 46: 344.
Kondo, Katsuhiko (1976) A cytotaxonomic study in some species of Drosera. Rhodora, 78: 532-541.
Kondo, K. and M. C. Olivier (1979) Chromosome numbers of four species of Drosera (Droseraceae). Annals of the Missouri Botanical Garden, 66: 584-587.
Kondo, K., and P. S. Lavarack. (1984) Study of some Australian species of Drosera L. (Droseraceae). Botanical Journal of the Linnean Society 88:317–333.
Sheikh, Shamimul Alam and Katsuhiko Kondo (1995) Differential Staining with Orcein, Giemsa, CMA, and DAPI for Comparative Chromosome Study of 12 Species of Australian Drosera (Droseraceae). American Journal of Botany 82(10):1278-1286. (jstor)
Sheikh, S. A., K. Kondo and Y. Hoshi (1995) Study on diffused centromeric nature of Drosera chromosomes. Cytologia 60: 43-47.
Kondo, Katsuhiko. Yoshikazu Hoshi, Takane Furut. (1998) Diffused Centromeric Chromosomes and Speciation in Drosera. Proc. 2nd Intl. Carniv. Pl. Conf. pages 4-5 ( PDF )
Hoshi, Yoshikazu and Katsuhiko Kondo (1998) Chromosome differentiation in Drosera, Subgenus Rorella, Section Rossolis. Cytologia 63: 199-211.
Hoshi, Yoshikazu and Katsuhiko Kondo (1998) A chromosome phylogeny of the Droseraceae by using CMA-DAPI fluorescent banding. Cytologia 63: 329-339.
Nontachaiyapoom, S., Inaga, S., Iino, A., and Kondo, K. (2000) Ultrastructure of chromatin organization in diffused-centromeric chromosomes of Drosera as compared with localized-centromeric chromosomes of Drosophyllum (Droseraceae) detected by scanning electron microscopy. Chromosome Science 4: 75-85.
Scanning electron microscope study of Drosera nuclei and chromosomes.
Hanson, Lynda, Kathryn A. McMahon, Margaret A. T. Johnson and Michael D. Bennett (2001) First Nuclear DNA C-values for 25 Angiosperm Families. Annals of Botany 87(2): 251-258. doi: 10.1006/anbo.2000.1325
Hanson, Lynda, Rebecca L. Brown, Amy Boyd, Margaret A. T. Johnson and Michael D. Bennett (2003) First Nuclear DNA C-values for 28 Angiosperm Genera. Annals of Botany 91(1): 31-38. doi: 10.1093/aob/mcg005
Hoshi, Yoshikazu (2002) Chromosome studies in Drosera (Droseraceae). Proc. 4th Intl. Carniv. Pl. Conf. pages 31-38 ( PDF )
A study of chromosomes in both mitotic and meiotic cells. All species showed similar pairing and movement in mitosis while Drosera petiolaris showed Drosera holocentric-style pairing and movement in the first meiotic division while Drosera rotundifolia showed pairing and movement reminiscent of single centromere chromosomes.
Rivadavia, Fernando, Katsuhiko Kondo, Masahiro Kato, and Mitsuyasu Hasebe (2003) Phylogeny of the sundews, Drosera (Droseraceae) based on chloroplast rbcL and nuclear 18S ribosomal DNA sequences. American Journal of Botany 90(1): 123-130. doi: 10.3732/ajb.90.1.123.
Massive phylogenetic study of Drosera.
Rivadavia, Fernando (2005) New Chromosome Numbers for Drosera L. (Droseraceae). Carniv. Pl. Newslett. 34(3):85-91 ( PDF )
Report and summary of Drosera chromosome numbers.
Lowrie, Allen and John G. Conran. (2007) A revision of the Drosera omissa/D. nitidula complex (Droseraceae) from south-west Western Australia. Taxon 56(2):533–544.
Kondo, K., and Nontachaiyapoom, S. (2008) An evidence on diffused centromeres in Drosera chromosomes provided by scanning electron microscopy. Chromosome Botany 3:79-81.
Visualization of a spindle fiber attachment.
Shirakawa, Junichi, Katsuya Nagano and Yoshikazu Hoshi (2011) A chromosome study of two centromere differentiating Drosera species, D. arcturi and D. regia. Caryologia 64(4): 453-463. (PDF from Caryologia)
Shirakawa, Junichi, Yoshikazu Hoshi and Katsuhiko Kondo (2011) Chromosome differentiation and genome organization in carnivorous plant family Droseraceae. Chromosome Botany 6: 111-119. doi: 10.3199/iscb.6.111
Shirakawa, Junichi, Yoshikazu Hoshi and Katsuhiko Kondo (2012) Polyploid Genome Structure of Drosera spatulata Complex (Droseraceae). Cytologia 77(1):97–106.
Detailed study of Drosera rotundifolia, Drosera spatulata (Japan), and Drosera tokaiensis chromosomes. The chromosomes are viewed with general DNA stains but more importantly also with 18s rDNA and genomic stains. The genomic stains allowed the viewing of Drosera rotundifolia and Drosera spatulata chromosomes in different colors in Drosera tokaiensis mitotic cells. The 18s rDNA stains indicated there has been genetic changes in Drosera tokaiensis compared to the parental species.
Rivadavia, F., V. F. O. de Miranda, G. Hoogenstrijd, F. Pinheiro, G. Heubl and A. Fleischmann. (2012) Is Drosera meristocaulis a pygmy sundew? Evidence of a long-distance dispersal between Western Australia and northern South America. Annals of Botany. doi: 10.1093/aob/mcs096
Heubl, G. and Wistuba, A. (1995) A cytological study of the genus Nepenthes L. (Nepenthaceae). Sendtnera 4:169-174.
A study of Nepenthes chromosomes. Proposes the base chromosome number for Droseraceae is x=5.
Lowrie, Allen (2013) Carnivorous Plants of Australia Magnum Opus. 3 volumes. Redfern Natural History Productions Ltd., Poole, GB.
Contains otherwise unpublished chromosome counts.
Drosera hybrids
Wood, C. E. (1955) Evidence for the hybrid origin of Drosera anglica. Rhodora 57: 105-130.
Sheridan, Philip (1987) A Preliminary Report on Drosera intermedia x D. capillaris. Carniv. Pl. Newslett. 16(3):71-73 ( PDF )
Drosera intermedia and Drosera capillaris have multiple forms in Florida. Now Drosera intermedia x D. capillaris hybrids have been found.
Schnell, Donald (1999) Drosera anglica Huds. vs. Drosera x anglica: What is the difference?. Carniv. Pl. Newslett. 28(4):107-115 ( PDF )
Comparison of characters delineating Drosera anglica and Drosera x anglica.
Snyder, Ivan (2003) Curious Natural Hybrid Sundews. Carniv. Pl. Newslett. 32(2):52-56 ( PDF )
A discussion of Drosera x obovata, Drosera anglica, and Drosera intermedia x D. capillaris along with crosses and colchicine treatments.
Brittnacher, John (2011) Drosera x hybrida rest in peace. Carniv. Pl. Newslett. 40(4):112-121 ( PDF )
History of Drosera x hybrida.
Kondo, K., AND M. Segawa. 1988. A cytotaxonomic study in artificial hybrids between Drosera anglica Huds. and its certain closely related species in series Drosera, section Drosera, subgenus Drosera, Drosera. La Kromosoma II 51-52: 1697–1709.
Nakano, M., E. Kinoshita and K. Ueda. (2000) Life history traits and coexistence of an amphidiploid, Drosera tokaiensis, and its parental species, D. rotundifolia and D. spatulata (Droseraceae). Plant Species Biology 19:59–72.
A study of the physical and life history traits of Drosera tokaiensis in relation to its parental species.
Allen Lowrie and John G. Conran (2007) Drosera × sidjamesii (Droseraceae): systematics and ecology of a natural hybrid from Western Australia. Australian Systematic Botany 20(1): 44–53. doi: 10.1071/SB04018
Schlauer, Jan (2010) Nomenclature of the Drosera anglica complex revisited. Carniv. Pl. Newslett. 39(2):46. ( PDF )
Appropriate taxonomic naming of Drosera anglica and its related hybrids if you treat Drosera anglica itself as a hybrid rather than an independent species.
Centromeres and Holocentric Chromosomes
Buscaino, Alessia, Robin Allshire and Alison Pidoux. (2010) Building centromeres: home sweet home or a nomadic existence? Current Opinion in Genetics & Development 20:1–9. doi: 10.1016/j.gde.2010.01.006.
How centromeres are constructed.
Dalal, Yamini, Takehito Furuyama, Danielle Vermaak, and Steven Henikoff. (2007) Structure, dynamics, and evolution of centromeric nucleosomes. Proceedings of the National Academy of Sciences 104(41):15974-15981. doi: 10.1073/pnas.0707648104.
A description of the structure and molecular biology of centromeres.
Gieni, Randall S., Gordon K.T. Chan, and Michael J. Hendzel. (2008) Epigenetics Regulate Centromere Formation and Kinetochore Function. Journal of Cellular Biochemistry 104:2027–2039. doi: 10.1002/jcb.21767.
A description of the structure and molecular biology of centromeres.
Roalson, E. H., A. G. McCubbin & R. Whitkus. (2007) Chromosome evolution in the Cyperales. In J. T. Columbus, E. A. Friar, J. M. Porter, L. M. Prince & M. G. Simpson (eds.), Monocots: comparative biology and evolution (Poales). Aliso 23:62–71.
A review of the karyotype evolution in the genus Carex (sedge) that has holocentric chromosomes. The genus has over 2000 species. Among the species measured, the haploid chromosome numbers range from n=6 to n=83 with evidence for both polyploidy and aneuploidy.
Hipp, Andrew L., Paul E. Rothrock, and Eric H. Roalson. (2009) The Evolution of Chromosome Arrangements in Carex (Cyperaceae). Botanical Review 75:96–109. doi: 10.1007/s12229-008-9022-8.
More than 100 Carex species exhibit as many as 10 cytotypes without being polyploid. Those cytotypes can to occur in a clinal fashion. Besides holocentric chromosomes the genus has a form of meiosis where sister chromatids separate in the first round and homologs separate in the second round and only produces one pollen grain from each pollen mother cell.
Chromosomes and Hybrids
Chapman, Mark A and Richard J. Abbott. (2010) Introgression of fitness genes across a ploidy barrier. New Phytologist 186:63–71. doi: 10.1111/j.1469-8137.2009.03091.x.
A study of the introgression of a flower character gene from the diploid Senecio squalidus to the tetraploid S. vulgaris.
Abbott, R. J., A. J. Lowe. (2004) Origins, establishment and evolution of new polyploid species: Senecio cambrensis and S. eboracensis in the British Isle. Biological Journal of the Linnean Society 82(4):467-474 doi: 10.1111/j.1095-8312.2004.00333.x.
Senecio cambrensis, a hexaploid arose multiple times via hybridization between between the native tetraploid and an introduced diploid. S. eboracensis is an introgressed tetraploid derived from the native tetraploid and an unreduced gamete from the triploid hybrid between the native tetraploid and introduced diploid.
Comai, L. (2005) The advantages and disadvantages of being polyploid. Nature Reviews Genetics 6(11):836–46. doi: 10.1038/nrg1711.
A good review of polyploidy in plants.
Soltis, D. E. and P. S. Soltis. (1999) Polyploidy: origins of species and genome evolution. Trends in Ecology and Evolution 14(9):349-351. doi: 10.1016/S0169-5347(99)01638-9.
Interbreeding between polyploids of independent origin can quickly contribute to the genetic variation of the polyploid species facilitating rapid evolution.
Buggs, RJA, P. S. Soltis, D. E. Soltis. (2009) Does hybridization between divergent progenitors drive whole-genome duplication? Molecular Ecology. Molecular Ecology 18:3334–3339. doi: 10.1111/j.1365-294X.2009.04285.x.
Parent species of homoploid hybrid species are more closely related then the parents of allopolyploid species.
Blanc G, Wolfe KH July (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16(7):1679–91. doi: 10.1105/tpc.021410.
The transcriptional profiles of genes duplicated during polyploidy can change dramatically during subsequent evolution.
Lim K.Y., D.E., Soltis, PS, Soltis, J. Tate, R. Matyasek, H. Srubarova, A. Kovarik, J.C. Pires, Z. Xiong, A.R. Leitch. (2008) Rapid Chromosome Evolution in Recently Formed Polyploids in Tragopogon (Asteraceae). PLoS ONE 3(10):e3353. doi: 10.1371/journal.pone.0003353.
Study of recent polyploids of 3 Eurasian diploids introduced in North America. The allotetraploid Tragopogon mirus recurred at least 13 times, T. miscellus possibly 21 times. The homoploid hybrid is highly sterile. The polyploids could not have existed more than 40 generations. Tetravalents were observed in meiotic cells and aneuploidy in mitotic cells. Minor chromosomal aberrations were observed.
Doyle, J. J., Doyle, J. L., Rausher, J. T. and Brown, A. H. D. (2004) Evolution of the perennial soybean polyploid complex (Glycine subgenus Glycine): a study of contrasts. Biological Journal of the Linnean Society 82 583–597. doi: 10.1111/j.1095-8312.2004.00343.x.
Review of polyploid complex in Australia consisting 6 of diploid genome groups and 9 resulting allopolyploids. Three genome groups present did not contribute to the polyploids. "Genome groups" is used because the formal taxonomy is not concordant with identified genomes. Some of the allopolyploids had multiple independent origins. It is hypothesized that polyploidization occurred as diploids colonized Australia 30,000 to 50,000 years ago.
Cronn, Richard and Jonathan F. Wendel (2004) Cryptic Trysts, Genomic Mergers, and Plant Speciation. New Phytologist 161(1):133-142. doi: 10.1046/j.1469-8137.2003.00947.x
Review of introgression between species and allopolyploidy speciation in Gossypium (cotton) including an allopolyploid lineage resulting from a relatively recent intercontinental dispersal event and introgression in a species resulting from an ancient intercontinental event. One fourth of the diploid Gossypium species they studied had evidence of introgression from genetically distant species.
Transposable Elements
Fedoroff, Nina V. (2012) Transposable Elements, Epigenetics, and Genome Evolution. Science 338(6108):758-767.
Review of the effects of transposable elements on genome evolution arguing that transposable elements are an important component of eukaryotic genomes and not parasites.
Feschotte, Cedric, Ning Jiang, and Susan R. Wessler. (2002) Plant transposable elements: where genetics meets genomics. Nature Reviews Genetics 3:329-341. doi: 10.1038/nrg793.
Review of the transposable element structure, function, and evolution in plants.
Wicker, Thomas, Francois Sabot, Aurelie Hua-Van, Jeffrey L. Bennetzen, Pierre Capy, Boulos Chalhoub, Andrew Flavell, Philippe Leroy, Michele Morgante, Olivier Panaud, Etienne Paux, Phillip SanMiguel and Alan H. Schulman. (2007) A unified classification system for eukaryotic transposable elements. Nature Reviews Genetics 8:973-982. doi: 10.1038/nrg2165
A review of transposable elements in eukaryotes and an attempt to classify them.
Vitte, Clementine, and Jeffrey L. Bennetzen (2006) Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proceedings of the National Academy of Science 103(47):17638–17643. doi: 10.1073/pnas.0605618103.
A study of five plant genomes looking at retrotransposon movement, age, and loss. Transposons were removed by unequal homologous recombination and illegitimate recombination.
Matzke, M., W. Gregor, M. F. Mette, W. Aufsatz, T. Kanno,J. Jakowitsch, A. J. M. Matzke. (2004) Endogenous pararetroviruses of polyploidy tobacco (Nicotiana tabacum) and its diploid progenitors, N. sylvestris and N. tomentosiformis. Biological Journal of the Linnean Society 82(4):627-638.
Infectious pararetroviruses are found integrated into genomes of many Nicotiana species. The integrated viruses are generally suppressed but can be activated in hybrids.
Laten, Howard M., Ericka R. Havecker, Lisa M. Farmer, and Daniel F. Voytas. (2003) SIRE1, an Endogenous Retrovirus Family from Glycine max, Is Highly Homogeneous and Evolutionarily Young. Mol. Biol. Evol. 20(8):1222–1230. doi: 10.1093/molbev/msg142.
Characterization of a retrovirus family in cotton. This retrotransposon contains the gene for a protein similar to the protein retroviruses use to encapsulate their RNA for transmission to a new host. It may be transmissible between plants via invertebrate vectors. There are about 1000 copies of SIRE1 in Glycine max.
Polyploids and Transposable Elements
Szadkowski, E., F. Eber, V. Huteau, M. Lode, C. Huneau, H. Belcra, O. Coriton, M. J. Manzanares-Dauleux, R. Delourme, G. J. King, B. Chalhoub, E. Jenczewski and A-M. Chevre. (2010) The first meiosis of resynthesized Brassica napus, a genome blender. New Phytologist 186:102–112. doi: 10.1111/j.1469-8137.2010.03182.x.
Synthetic allotetraploid Brassica napus showed chromosome rearrangements in the first generation meiotic cells. Cytoplasmic background influenced the frequency of aberrations.
Petit, M., C. Guidat, J. Daniel, E. Denis, E. Montoriol, Q. T. Bui, K. Y. Lim, A. Kovarik, A. R. Leitch, M-A. Grandbastien and C. Mhiri. (2010) Mobilization of retrotransposons in synthetic allotetraploid tobacco. New Phytologist 186:135–147. doi: 10.1111/j.1469-8137.2009.03140.x.
Documented retrotransposon amplification in newly synthesized allotetraploid Nicotiana tabacum. There were also losses in retrotransposons at particular sites even though retrotransposons can not excise. Most of the changes occurred in only one of the parent genomes, possibly from failed chromosome pairing.
Martienssen, Robert A. (2010) Heterochromatin, small RNA and post-fertilization dysgenesis in allopolyploid and interploid hybrids of Arabidopsis. New Phytologist 186:46–53. doi: 10.1111/j.1469-8137.2010.03193.x.
Hybrid seed failure can result from a genome imbalance between the parents and from the presence or lack of factors silencing transposable elements.
Adams K. L. and J. F. Wendel (2005) Polyploidy and genome evolution in plants. Current Opinion in Plant Biology 8(2): 135–41. doi: 10.1016/j.pbi.2005.01.001.
Reviews many studies demonstrating cycles of genome doubling via polyploidy followed by genome trimming.
Parisod, Christian, Rolf Holderegger and Christian Brochmann. (2009) Evolutionary consequences of autopolyploidy. New Phytologist 186:5–17. doi: 10.1111/j.1469-8137.2009.03142.x
Autopolyploidy does not trigger the immediate major genome restructuring seen in allopolyploids although over longer time frames restructuring does occur. Autopolyploids tend to be found in disturbed habitats where they may have a short term advantage over the diploid progenitor.
Ancient Polyploid Events
Paterson, A. H., Bowers, J. E., Chapman, B. A. (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proceedings of the National Academy of Sciences 101(26):9903–9908 . doi: 10.1073/pnas.0307901101.
Uses genome analysis to trace chromosomal elements resulting from a polyploidization event that occurred prior to the divergence of Sorghum and Oryza (rice). Oryza is more strongly diploidized than Sorghum showing the loss of many chromosomal segments.
Bowers, J. E., Chapman, B. A., Rong, J., Paterson, A. H. (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422(6930):433–438. doi:10.1038/nature01521.
Genome analysis of Arabidopsis thaliana delimiting retained and lost sequences from an ancient polyploidization event.
Lysak, Martin A., Alexandre Berr, Ales Pecinka, Renate Schmidt, Kim McBreen, and Ingo Schubert. (2006) Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proceedings of the National Academy of Sciences 103(13):5224–5229. doi: 10.1073/pnas.0510791103.
An analysis of the chromosomes of Arabidopsis thaliana and 5 related species that evolved after a polyploidization event showed that two species retained the original n=8 chromosomes while the four other species experienced chromosome number decreases to n=5, 6, and 7 via chromosome fusions, translocations, and losses.
Cui L., P. K. Wall, J. H. Leebens-Mack, B. G. Lindsay, D. E. Soltis, J. J. Doyle, P. S. Soltis, J. E. Carlson, K. Arumuganathan, A. Barakat, V. A. Albert, H. Ma, and C. W. dePamphilis. (2006) Widespread genome duplications throughout the history of flowering plants. Genome Research 16(6):738–749. doi: 10.1101/gr.4825606.
Used the number of mutations in duplicated genes to estimate number of past polyploidy events in various lineages.
Putnam NH, Butts T, Ferrier DE, et al. (2008) The amphioxus genome and the evolution of the chordate karyotype. Nature 453(7198):1064–1071. doi: 10.1038/nature06967.
A comparison of the amphioxus genome with the human genome.
Blanc, G., Wolfe K. H. (2000) Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16(7):1667–1678. doi: 10.1105/tpc.021345.
Used the number of mutations in duplicated genes to estimate dates of past polyploidy events.
Doyle, Jeff J. and Ashley N. Egan. (2010) Dating the origins of polyploidy events. New Phytologist 186:73–85. doi: 10.1111/j.1469-8137.2009.03118.x.
Because of the mode of origin of polyploids and that genetic comparisons are with extant parents that have evolved since the polyploidization event it is difficult if not impossible to put dates on the origin using simple DNA and other genetic analysis.